AlphaFold: Deep Learning Marked 2020 With A Life-Impacting Breakthrough
A breakthrough stunned world-leading scientists at the close of 2020.
“But when I first saw these results, I nearly fell off my chair.”
“…a once in a generation advance.”
“I was beginning to think it would not get solved in my lifetime.”
“It has occurred decades before many people in the field would have predicted.”
Even the staid technology journal, IEEE Spectrum, lost restraint. “AlphaFold’s discovery is right up there with the likes of James Watson and Francis Crick’s DNA double-helix model.”
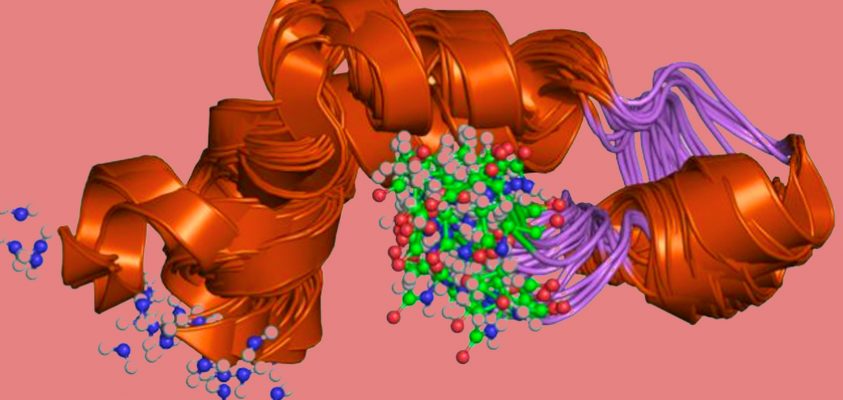
What happened? Alphabet subsidiary, DeepMind, cracked a monumental problem in computing, AI, and biology: protein folding. It is finding the three-dimensional shape of a protein. DeepMind’s program, AlphaFold, does it as well as the best physical laboratory experiments. What can take years and millions of dollars of investment in a lab, AlphaFold does over a weekend for the cost of computer time.
It is another point in the problem-busting tally racked up by artificial intelligence (AI) across diverse fields in the last 10 years.
Why Solving Folding Is So Important
The way a protein folds is essential for life and inconceivably difficult to calculate. Figuring out exactly how proteins fold is key to understanding what allows life, as we know it, to exist.
Most of what goes on in your body’s cells, like growing, taking in nutrition, and fighting off disease, happens using proteins.
Proteins are long chains of organic molecules called amino acids. We humans have 80,000 to 400,000different types of protein keeping our bodies working. They vary in length from 30 amino acids up to 35,000. Proteins twist and fold themselves into complex shapes, like a piece of string that you’ve rolled into a ball between your hands. That tangled 3D shape determines what a protein can do in your body.
University of Maryland Professor John Moult, whose 26-year bi-annual contest sparked the breakthrough, explained. “We have been stuck on this one problem – how do proteins fold up – for nearly 50 years.“

Astronomically Complex
There are more ways for a protein to fold up than atoms in the universe (10300 vs. 1080). Yet each protein folds itself just one way, the same way, virtually every time.

Imagine thousands of assorted Lego pieces snapping themselves together into the same model, time after time. Change one piece, and the whole model rearranges into something different.
For months, even years, scientists work in the lab to find the shape of a given protein. They use specialized multi-million-dollar equipment. As for computer simulations, the eminent molecular biologist, Cyrus Levinthal, estimated that it would take longer than the universe’s age (13.8 billion years) to find a protein’s shape by calculating the interactions between all of its atoms.
With AlphaFold, a researcher can correctly fold a protein in a couple of days.
What AlphaFold Means For Medicine
This advance may significantly accelerate drug development and slash R&D costs.
Knowing a protein’s folded shape is like having the key to a lock. Much of the action in biology happens when a molecule fits neatly into a feature on a protein. For example:

Drugs work by fitting to proteins and altering their function.
Professor Moult and other experts believe that computer simulations will speed drug creation. They could swiftly calculate which drugs are likely to bind well to particular viruses, bacteria, or human proteins.
Saving significant time and money drops the investment hurdle for drug development. Many more researchers and entrepreneurs will explore a vastly greater number of therapies for multiple diseases. When small teams, spinning up servers at AWS, can compete with massive pharma companies, the drug industry’s shape will be revolutionized.
How Did AlphaFold Happen?
How did the DeepMind team do it? Their method, not yet fully published, leaned heavily on the usual AI suspect: Deep Learning.
Deep Learning is a pattern-matching savant. After training a Deep Learning system with many examples like “This is a picture of a cat,” it will tell you whether subsequent pictures are cats. In training the Deep Learning piece of AlphaFold, the Deep Mind team fed it 170,000 examples of amino acid chains and the resulting folded proteins. After training, when handed a new chain, it created a new 3D folded protein model mimicking patterns it had seen before.
This AI does not think like our human brain. It is different but radically better in one very narrow task.
Part Of A Pattern
AlphaFold’s success is itself part of a pattern. Researchers wielding Deep Learning mastered many seemingly intractable computing problems in the last decade:
- 2012: it was used to crush the ImageNet picture identification contest.
- 2016: the method conquered Go, the ancient Chinese game so complex that, like protein folding, it has more possibilities (~10360) than there are atoms in the universe.
- 2017: Deep Learning was at the controls of self-driving taxis, “Robotaxis,” that hit the streets in significant numbers.
- 2018: Deep Learning is also behind the speech recognition in your smartphone and smart speakers like Amazon’s Alexa.
Abundant Potential
David Baker, head of the Institute for Protein Design at the University of Washington, says of AlphaFold, “Once they describe to the world how they do it then a thousand flowers will bloom… People will be using it for all kinds of different things, things that we can’t imagine now.”
Janet Thornton, Director Emeritus of the European Bioinformatics Institute, notes, “It is the first real step toward… building proteins that fulfill a specific function. From protein therapeutics to biofuels or enzymes that eat plastic, the possibilities are endless.”
Exiting 2020, a year overflowing with pain and frustration, I share their hope for abundant discoveries. Thousands of teams running hundreds of millions of cheap digital experiments around drugs, energy, waste, and more, will reshape many of our biggest challenges.
Cover Image: Jawahar Swaminathan and MSD staff at the European Bioinformatics Institute